import pymc as pm
import pandas as pd
import numpy as np
from pytensor import tensor as pt
import arviz as az
from matplotlib import pyplot as plt
import seaborn as sns
from pymc import do, observeMeasurment Model
df = pd.read_csv('sem_data.csv')
df.head()| motiv | harm | stabi | ppsych | ses | verbal | read | arith | spell | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | -7.907122 | -5.075312 | -3.138836 | -17.800210 | 4.766450 | -3.633360 | -3.488981 | -9.989121 | -6.567873 |
| 1 | 1.751478 | -4.155847 | 3.520752 | 7.009367 | -6.048681 | -7.693461 | -4.520552 | 8.196238 | 8.778973 |
| 2 | 14.472570 | -4.540677 | 4.070600 | 23.734260 | -16.970670 | -3.909941 | -4.818170 | 7.529984 | -5.688716 |
| 3 | -1.165421 | -5.668406 | 2.600437 | 1.493158 | 1.396363 | 21.409450 | -3.138441 | 5.730547 | -2.915676 |
| 4 | -4.222899 | -10.072150 | -6.030737 | -5.985864 | -18.376400 | -1.438816 | -2.009742 | -0.623953 | -1.024624 |
coords = {
"obs": list(range(len(df))),
"indicators": ["motiv", "harm", "stabi", "verbal", "ses", "ppsych", "read", "arith", "spell"],
"indicators_1": ["motiv", "harm", "stabi"],
"indicators_2": ["verbal", "ses", "ppsych"],
"indicators_3": ["read", "arith", "spell"],
"latent": ["adjust", "risk", "achieve"],
}
def make_lambda(indicators, name='lambdas1', priors=[1, 10]):
""" Takes an argument indicators which is a string in the coords dict"""
temp_name = name + '_'
lambdas_ = pm.Normal(temp_name, priors[0], priors[1], dims=(indicators))
# Force a fixed scale on the factor loadings for factor 1
lambdas_1 = pm.Deterministic(
name, pt.set_subtensor(lambdas_[0], 1), dims=(indicators)
)
return lambdas_1obs_idx = list(range(len(df)))
with pm.Model(coords=coords) as model:
# Set up Factor Loadings
lambdas_1 = make_lambda('indicators_1', 'lambdas1')
lambdas_2 = make_lambda('indicators_2', 'lambdas2', priors=[1, 2])
lambdas_3 = make_lambda('indicators_3', 'lambdas3')
# Specify covariance structure between latent factors
kappa = 0
sd_dist = pm.Exponential.dist(1.0, shape=3)
chol, _, _ = pm.LKJCholeskyCov("chol_cov", n=3, eta=2, sd_dist=sd_dist, compute_corr=True)
ksi = pm.MvNormal("ksi", kappa, chol=chol, dims=("obs", "latent"))
# Construct Pseudo Observation matrix based on Factor Loadings
m1 = ksi[obs_idx, 0] * lambdas_1[0]
m2 = ksi[obs_idx, 0] * lambdas_1[1]
m3 = ksi[obs_idx, 0] * lambdas_1[2]
m4 = ksi[obs_idx, 1] * lambdas_2[0]
m5 = ksi[obs_idx, 1] * lambdas_2[1]
m6 = ksi[obs_idx, 1] * lambdas_2[2]
m7 = ksi[obs_idx, 2] * lambdas_3[0]
m8 = ksi[obs_idx, 2] * lambdas_3[1]
m9 = ksi[obs_idx, 2] * lambdas_3[2]
mu = pm.Deterministic("mu", pm.math.stack([m1, m2, m3, m4, m5, m6, m7, m8, m9]).T)
## Error Terms
Psi = pm.InverseGamma("Psi", 2, 13, dims="indicators")
# Likelihood
_ = pm.Normal(
"likelihood",
mu,
Psi,
observed=df[coords['indicators']].values,
)
idata = pm.sample(
draws=1000,
chains=4,
nuts_sampler="numpyro", target_accept=0.95, idata_kwargs={"log_likelihood": True},
tune=2000,
random_seed=150
)
idata.extend(pm.sample_posterior_predictive(idata))
pm.model_to_graphviz(model)--------------------------------------------------------------------------- KeyboardInterrupt Traceback (most recent call last) Cell In[76], line 38 30 # Likelihood 31 _ = pm.Normal( 32 "likelihood", 33 mu, 34 Psi, 35 observed=df[coords['indicators']].values, 36 ) ---> 38 idata = pm.sample( 39 draws=1000, 40 chains=4, 41 nuts_sampler="numpyro", target_accept=0.95, idata_kwargs={"log_likelihood": True}, 42 tune=2000, 43 random_seed=150 44 ) 45 idata.extend(pm.sample_posterior_predictive(idata)) 47 pm.model_to_graphviz(model) File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/pymc/sampling/mcmc.py:727, in sample(draws, tune, chains, cores, random_seed, progressbar, progressbar_theme, step, var_names, nuts_sampler, initvals, init, jitter_max_retries, n_init, trace, discard_tuned_samples, compute_convergence_checks, keep_warning_stat, return_inferencedata, idata_kwargs, nuts_sampler_kwargs, callback, mp_ctx, blas_cores, model, **kwargs) 722 raise ValueError( 723 "Model can not be sampled with NUTS alone. Your model is probably not continuous." 724 ) 726 with joined_blas_limiter(): --> 727 return _sample_external_nuts( 728 sampler=nuts_sampler, 729 draws=draws, 730 tune=tune, 731 chains=chains, 732 target_accept=kwargs.pop("nuts", {}).get("target_accept", 0.8), 733 random_seed=random_seed, 734 initvals=initvals, 735 model=model, 736 var_names=var_names, 737 progressbar=progressbar, 738 idata_kwargs=idata_kwargs, 739 compute_convergence_checks=compute_convergence_checks, 740 nuts_sampler_kwargs=nuts_sampler_kwargs, 741 **kwargs, 742 ) 744 if isinstance(step, list): 745 step = CompoundStep(step) File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/pymc/sampling/mcmc.py:356, in _sample_external_nuts(sampler, draws, tune, chains, target_accept, random_seed, initvals, model, var_names, progressbar, idata_kwargs, compute_convergence_checks, nuts_sampler_kwargs, **kwargs) 353 elif sampler in ("numpyro", "blackjax"): 354 import pymc.sampling.jax as pymc_jax --> 356 idata = pymc_jax.sample_jax_nuts( 357 draws=draws, 358 tune=tune, 359 chains=chains, 360 target_accept=target_accept, 361 random_seed=random_seed, 362 initvals=initvals, 363 model=model, 364 var_names=var_names, 365 progressbar=progressbar, 366 nuts_sampler=sampler, 367 idata_kwargs=idata_kwargs, 368 compute_convergence_checks=compute_convergence_checks, 369 **nuts_sampler_kwargs, 370 ) 371 return idata 373 else: File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/pymc/sampling/jax.py:615, in sample_jax_nuts(draws, tune, chains, target_accept, random_seed, initvals, jitter, model, var_names, nuts_kwargs, progressbar, keep_untransformed, chain_method, postprocessing_backend, postprocessing_vectorize, postprocessing_chunks, idata_kwargs, compute_convergence_checks, nuts_sampler) 612 raise ValueError(f"{nuts_sampler=} not recognized") 614 tic1 = datetime.now() --> 615 raw_mcmc_samples, sample_stats, library = sampler_fn( 616 model=model, 617 target_accept=target_accept, 618 tune=tune, 619 draws=draws, 620 chains=chains, 621 chain_method=chain_method, 622 progressbar=progressbar, 623 random_seed=random_seed, 624 initial_points=initial_points, 625 nuts_kwargs=nuts_kwargs, 626 ) 627 tic2 = datetime.now() 629 if idata_kwargs is None: File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/pymc/sampling/jax.py:469, in _sample_numpyro_nuts(model, target_accept, tune, draws, chains, chain_method, progressbar, random_seed, initial_points, nuts_kwargs) 455 pmap_numpyro.run( 456 map_seed, 457 init_params=initial_points, (...) 465 ), 466 ) 468 raw_mcmc_samples = pmap_numpyro.get_samples(group_by_chain=True) --> 469 sample_stats = _numpyro_stats_to_dict(pmap_numpyro) 470 return raw_mcmc_samples, sample_stats, numpyro File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/pymc/sampling/jax.py:409, in _numpyro_stats_to_dict(posterior) 407 data[name] = value 408 if stat == "num_steps": --> 409 data["tree_depth"] = np.log2(value).astype(int) + 1 410 return data File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/jax/_src/array.py:425, in ArrayImpl.__array__(self, dtype, context, copy) 422 def __array__(self, dtype=None, context=None, copy=None): 423 # copy argument is supported by np.asarray starting in numpy 2.0 424 kwds = {} if copy is None else {'copy': copy} --> 425 return np.asarray(self._value, dtype=dtype, **kwds) File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/jax/_src/profiler.py:333, in annotate_function.<locals>.wrapper(*args, **kwargs) 330 @wraps(func) 331 def wrapper(*args, **kwargs): 332 with TraceAnnotation(name, **decorator_kwargs): --> 333 return func(*args, **kwargs) 334 return wrapper File ~/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/jax/_src/array.py:645, in ArrayImpl._value(self) 643 npy_value = np.empty(self.shape, self.dtype) 644 for i, ind in _cached_index_calc(self.sharding, self.shape): --> 645 npy_value[ind] = self._arrays[i]._single_device_array_to_np_array() 646 self._npy_value = npy_value 647 self._npy_value.flags.writeable = False KeyboardInterrupt:
az.summary(idata, var_names=["lambdas1", "lambdas2", "lambdas3"])/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| lambdas1[motiv] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 4000.0 | 4000.0 | NaN |
| lambdas1[harm] | 0.895 | 0.043 | 0.814 | 0.973 | 0.002 | 0.001 | 521.0 | 1401.0 | 1.01 |
| lambdas1[stabi] | 0.703 | 0.047 | 0.617 | 0.791 | 0.002 | 0.001 | 801.0 | 1571.0 | 1.01 |
| lambdas2[verbal] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 4000.0 | 4000.0 | NaN |
| lambdas2[ses] | 0.842 | 0.087 | 0.679 | 1.000 | 0.004 | 0.003 | 453.0 | 1294.0 | 1.01 |
| lambdas2[ppsych] | -0.801 | 0.084 | -0.969 | -0.653 | 0.004 | 0.003 | 521.0 | 1276.0 | 1.00 |
| lambdas3[read] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 4000.0 | 4000.0 | NaN |
| lambdas3[arith] | 0.841 | 0.035 | 0.773 | 0.906 | 0.001 | 0.001 | 1955.0 | 2586.0 | 1.00 |
| lambdas3[spell] | 0.981 | 0.029 | 0.927 | 1.037 | 0.001 | 0.001 | 1123.0 | 1700.0 | 1.00 |
az.plot_trace(idata, var_names=['lambdas1', 'lambdas2', 'lambdas3'])
plt.tight_layout();/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/density_utils.py:488: UserWarning: Your data appears to have a single value or no finite values
warnings.warn("Your data appears to have a single value or no finite values")
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/density_utils.py:488: UserWarning: Your data appears to have a single value or no finite values
warnings.warn("Your data appears to have a single value or no finite values")
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/density_utils.py:488: UserWarning: Your data appears to have a single value or no finite values
warnings.warn("Your data appears to have a single value or no finite values")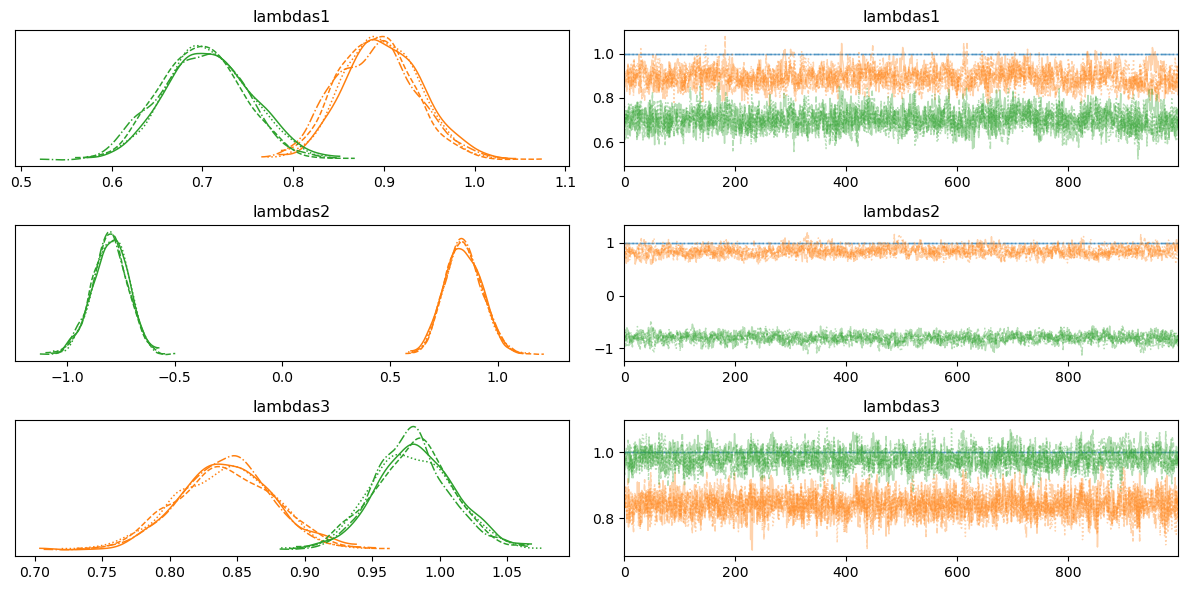
def make_factor_loadings_df(idata):
factor_loadings = pd.DataFrame(
az.summary(
idata, var_names=["lambdas1", "lambdas2", "lambdas3"]
)["mean"]
).reset_index()
factor_loadings["factor"] = factor_loadings["index"].str.split("[", expand=True)[0]
factor_loadings.columns = ["factor_loading", "factor_loading_weight", "factor"]
factor_loadings["factor_loading_weight_sq"] = factor_loadings["factor_loading_weight"] ** 2
factor_loadings["sum_sq_loadings"] = factor_loadings.groupby("factor")[
"factor_loading_weight_sq"
].transform(sum)
factor_loadings["error_variances"] = az.summary(idata, var_names=["Psi"])["mean"].values
factor_loadings["total_indicator_variance"] = (
factor_loadings["factor_loading_weight_sq"] + factor_loadings["error_variances"]
)
factor_loadings["total_variance"] = factor_loadings["total_indicator_variance"].sum()
factor_loadings["indicator_explained_variance"] = (
factor_loadings["factor_loading_weight_sq"] / factor_loadings["total_variance"]
)
factor_loadings["factor_explained_variance"] = (
factor_loadings["sum_sq_loadings"] / factor_loadings["total_variance"]
)
num_cols = [c for c in factor_loadings.columns if not c in ["factor_loading", "factor"]]
return factor_loadings
pd.set_option("display.max_colwidth", 15)
factor_loadings = make_factor_loadings_df(idata)
num_cols = [c for c in factor_loadings.columns if not c in ["factor_loading", "factor"]]
factor_loadings.style.format("{:.3f}", subset=num_cols).background_gradient(
axis=0, subset=["indicator_explained_variance", "factor_explained_variance"]
)/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/var/folders/__/ng_3_9pn1f11ftyml_qr69vh0000gn/T/ipykernel_52977/2929805399.py:12: FutureWarning: The provided callable <built-in function sum> is currently using SeriesGroupBy.sum. In a future version of pandas, the provided callable will be used directly. To keep current behavior pass the string "sum" instead.
].transform(sum)| factor_loading | factor_loading_weight | factor | factor_loading_weight_sq | sum_sq_loadings | error_variances | total_indicator_variance | total_variance | indicator_explained_variance | factor_explained_variance | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | lambdas1[motiv] | 1.000 | lambdas1 | 1.000 | 2.295 | 3.676 | 4.676 | 60.931 | 0.016 | 0.038 |
| 1 | lambdas1[harm] | 0.895 | lambdas1 | 0.801 | 2.295 | 5.623 | 6.424 | 60.931 | 0.013 | 0.038 |
| 2 | lambdas1[stabi] | 0.703 | lambdas1 | 0.494 | 2.295 | 7.605 | 8.099 | 60.931 | 0.008 | 0.038 |
| 3 | lambdas2[verbal] | 1.000 | lambdas2 | 1.000 | 2.351 | 6.915 | 7.915 | 60.931 | 0.016 | 0.039 |
| 4 | lambdas2[ses] | 0.842 | lambdas2 | 0.709 | 2.351 | 8.027 | 8.736 | 60.931 | 0.012 | 0.039 |
| 5 | lambdas2[ppsych] | -0.801 | lambdas2 | 0.642 | 2.351 | 8.242 | 8.884 | 60.931 | 0.011 | 0.039 |
| 6 | lambdas3[read] | 1.000 | lambdas3 | 1.000 | 2.670 | 3.409 | 4.409 | 60.931 | 0.016 | 0.044 |
| 7 | lambdas3[arith] | 0.841 | lambdas3 | 0.707 | 2.670 | 6.168 | 6.875 | 60.931 | 0.012 | 0.044 |
| 8 | lambdas3[spell] | 0.981 | lambdas3 | 0.962 | 2.670 | 3.951 | 4.913 | 60.931 | 0.016 | 0.044 |
fig, axs = plt.subplots(1, 2, figsize=(20, 9))
axs = axs.flatten()
az.plot_energy(idata, ax=axs[0])
az.plot_forest(idata, var_names=["lambdas1", "lambdas2", "lambdas3"], combined=True, ax=axs[1]);
plt.tight_layout();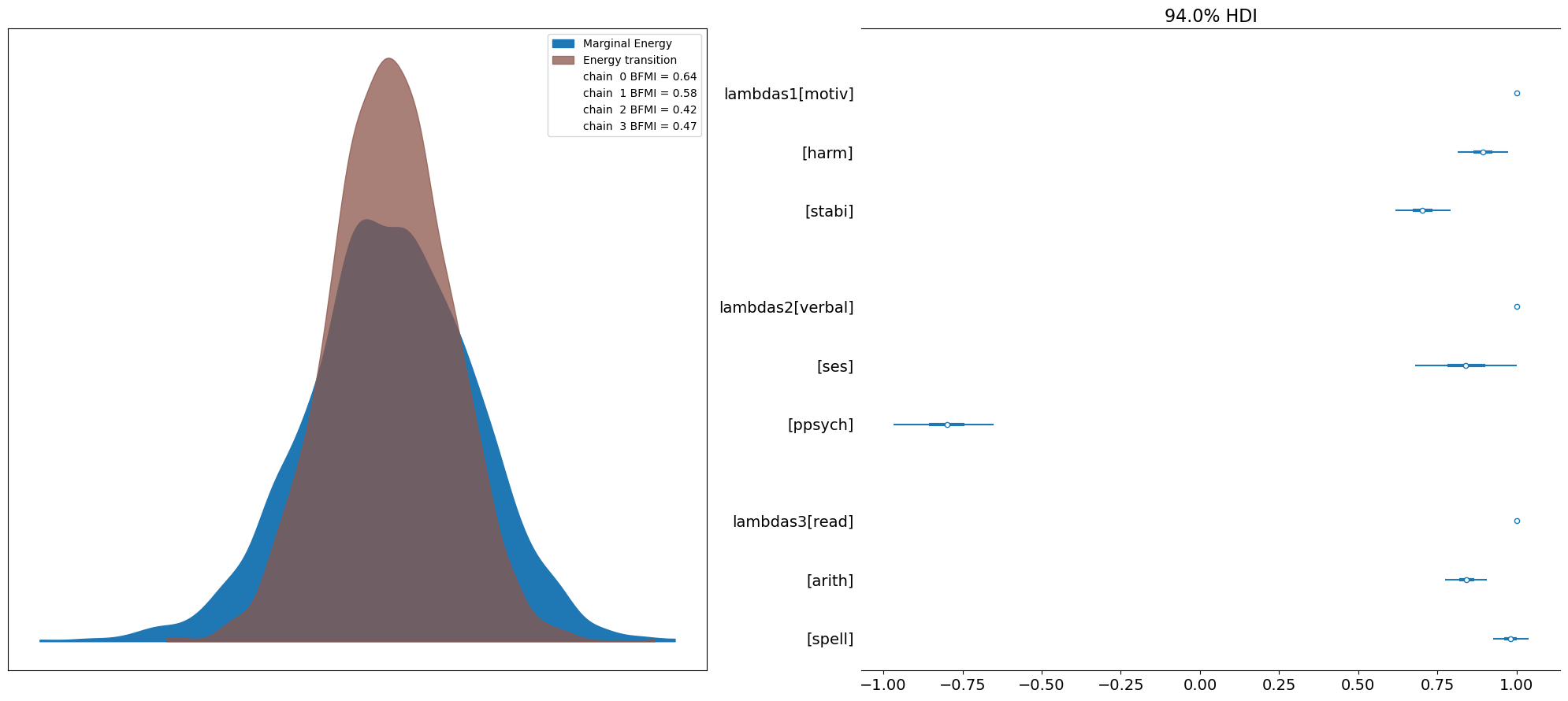
drivers = coords['indicators']
def get_posterior_resids(idata, samples=100, metric="cov"):
resids = []
for i in range(100):
if metric == "cov":
model_cov = pd.DataFrame(
az.extract(idata["posterior_predictive"])["likelihood"][:, :, i]
).cov()
obs_cov = df[drivers].cov()
else:
model_cov = pd.DataFrame(
az.extract(idata["posterior_predictive"])["likelihood"][:, :, i]
).corr()
obs_cov = df[drivers].corr()
model_cov.index = obs_cov.index
model_cov.columns = obs_cov.columns
residuals = model_cov - obs_cov
resids.append(residuals.values.flatten())
residuals_posterior = pd.DataFrame(pd.DataFrame(resids).mean().values.reshape(9, 9))
residuals_posterior.index = obs_cov.index
residuals_posterior.columns = obs_cov.index
return residuals_posterior
residuals_posterior_cov = get_posterior_resids(idata, 1000)
residuals_posterior_corr = get_posterior_resids(idata, 1000, metric="corr")fig, ax = plt.subplots(figsize=(20, 10))
mask = np.triu(np.ones_like(residuals_posterior_corr, dtype=bool))
ax = sns.heatmap(residuals_posterior_corr, annot=True, cmap="bwr", mask=mask)
ax.set_title("Residuals between Model Implied and Sample Correlations \n CFA", fontsize=25);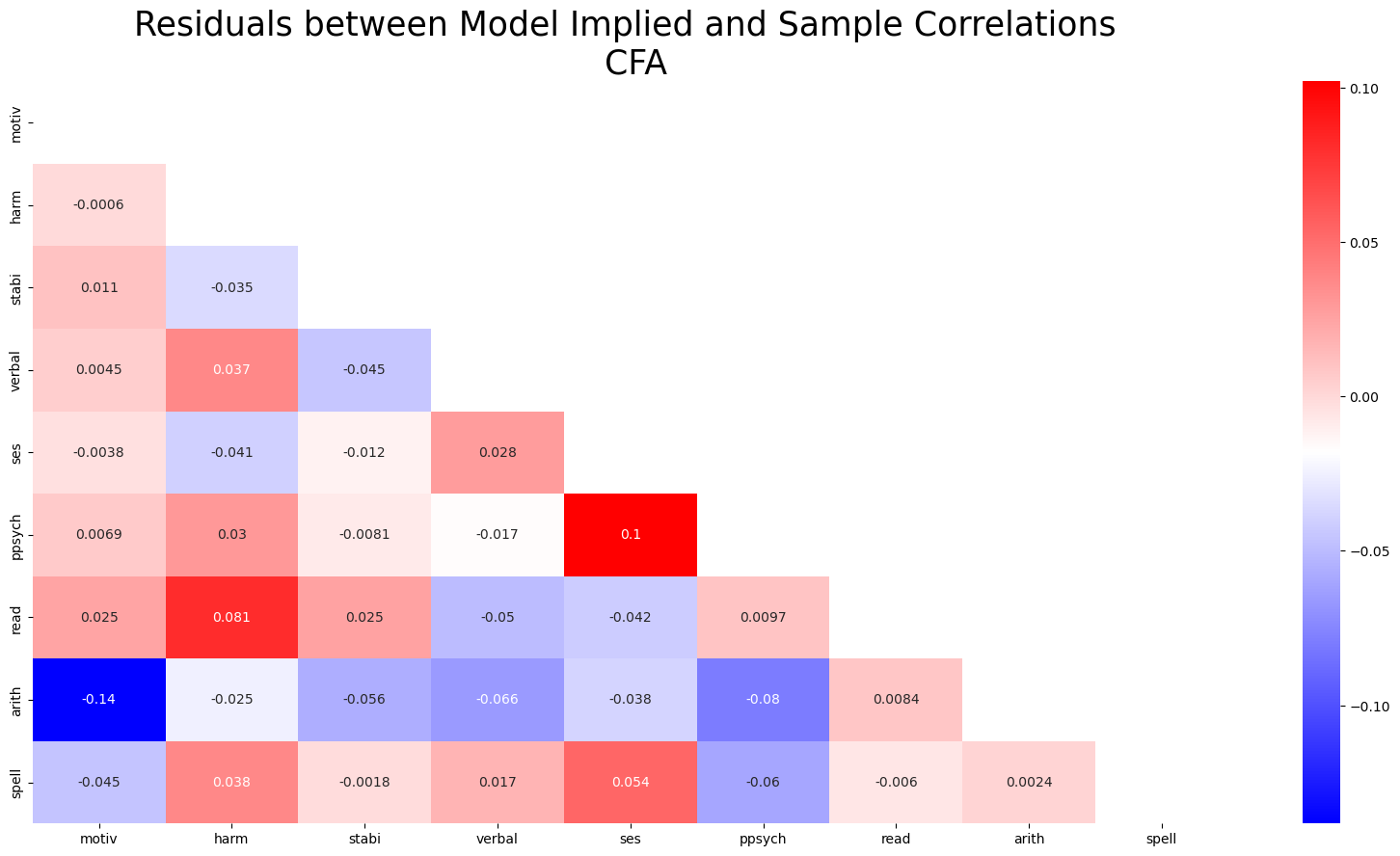
def make_ppc(
idata,
samples=100,
drivers=drivers,
dims=(3, 3),
):
fig, axs = plt.subplots(dims[0], dims[1], figsize=(20, 10))
axs = axs.flatten()
for i in range(len(drivers)):
for j in range(samples):
temp = az.extract(idata["posterior_predictive"].sel({"likelihood_dim_3": i}))[
"likelihood"
].values[:, j]
temp = pd.DataFrame(temp, columns=["likelihood"])
if j == 0:
axs[i].hist(df[drivers[i]], alpha=0.3, ec="black", bins=20, label="Observed Scores")
axs[i].hist(
temp["likelihood"], color="purple", alpha=0.1, bins=20, label="Predicted Scores"
)
else:
axs[i].hist(df[drivers[i]], alpha=0.3, ec="black", bins=20)
axs[i].hist(temp["likelihood"], color="purple", alpha=0.1, bins=20)
axs[i].set_title(f"Posterior Predictive Checks {drivers[i]}")
axs[i].legend()
plt.tight_layout()
plt.show()
make_ppc(idata)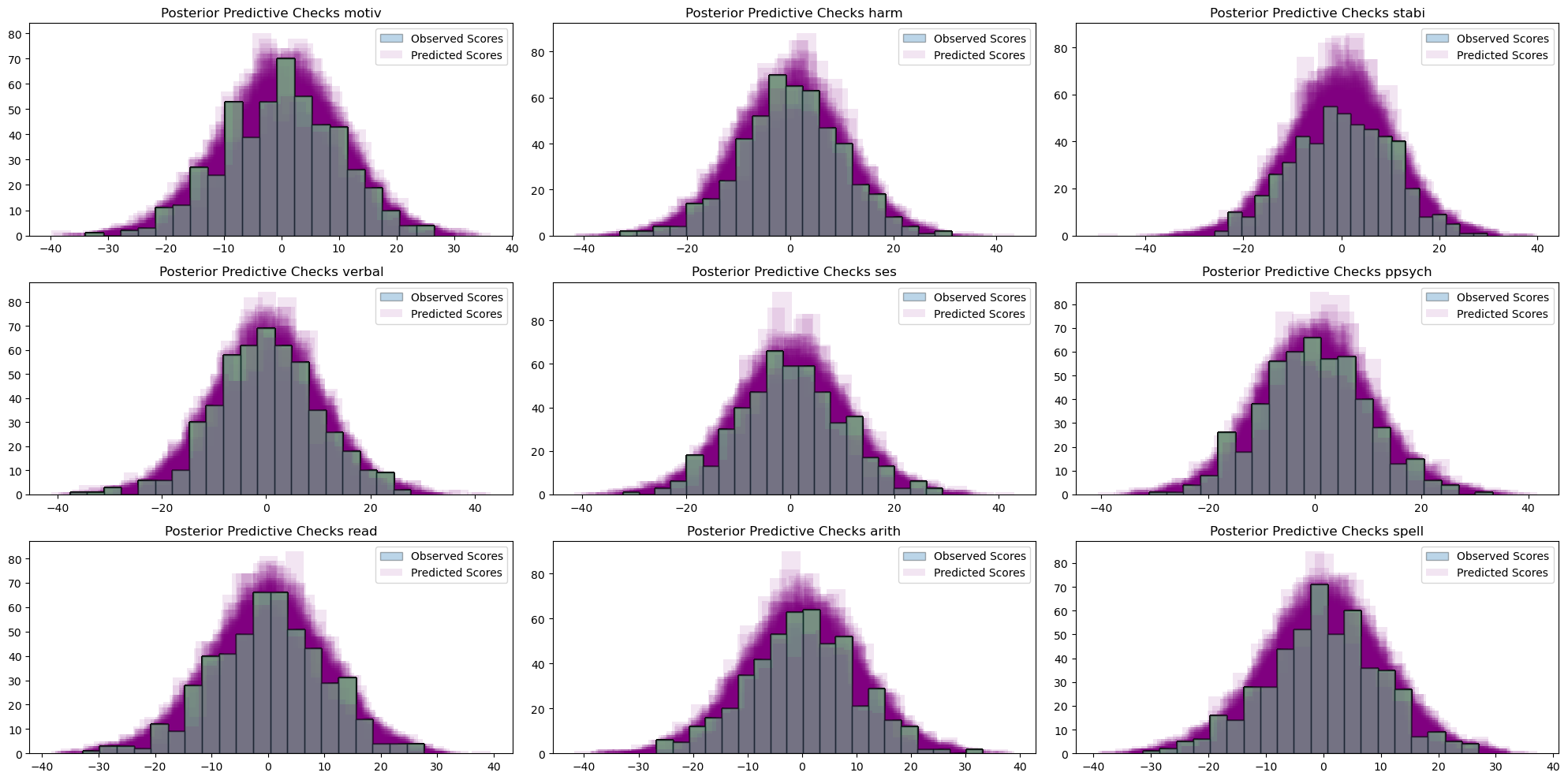
SEM Model
Making the Structural Matrix
import numpy as np
import pandas as pd
data = np.random.multivariate_normal([0,0,0], [[1, 0, 0], [0,1, 0], [0, 0, 1]], size=(500))
data = np.ones((500, 3))
betas = np.zeros((3, 3))
betas[1, 0] = 0.5
betas[1, 2] = 0.5
betas[0, 2] = 0.7
B_matrix = pd.DataFrame(betas, index=['adjust_coef', 'risk_coef', 'achieve_coef'],
columns=['adjust_target', 'risk_target', 'achieve_target'])
data = pd.DataFrame(data, columns=['adjust', 'risk', 'achieve'])
B_matrix| adjust_target | risk_target | achieve_target | |
|---|---|---|---|
| adjust_coef | 0.0 | 0.0 | 0.7 |
| risk_coef | 0.5 | 0.0 | 0.5 |
| achieve_coef | 0.0 | 0.0 | 0.0 |
data.head()| adjust | risk | achieve | |
|---|---|---|---|
| 0 | 1.0 | 1.0 | 1.0 |
| 1 | 1.0 | 1.0 | 1.0 |
| 2 | 1.0 | 1.0 | 1.0 |
| 3 | 1.0 | 1.0 | 1.0 |
| 4 | 1.0 | 1.0 | 1.0 |
np.dot(data, B_matrix)array([[0.5, 0. , 1.2],
[0.5, 0. , 1.2],
[0.5, 0. , 1.2],
...,
[0.5, 0. , 1.2],
[0.5, 0. , 1.2],
[0.5, 0. , 1.2]])Fitting the Model
def make_sem():
coords = {
"obs": list(range(len(df))),
"indicators": ["motiv", "harm", "stabi", "verbal", "ses", "ppsych", "read", "arith", "spell"],
"indicators_1": ["motiv", "harm", "stabi"],
"indicators_2": ["verbal", "ses", "ppsych"],
"indicators_3": ["read", "arith", "spell"],
"latent": ["adjust", "risk", "achieve"],
"paths": ["risk->adjust", "risk-->achieve", "adjust-->achieve"]
}
obs_idx = list(range(len(df)))
with pm.Model(coords=coords) as model:
# Set up Factor Loadings
lambdas_1 = make_lambda('indicators_1', 'lambdas1', priors=[1, .5]) #adjust
lambdas_2 = make_lambda('indicators_2', 'lambdas2', priors=[1, .5]) # risk
lambdas_3 = make_lambda('indicators_3', 'lambdas3', priors=[1, .5]) # achieve
# Specify covariance structure between latent factors
sd_dist = pm.Exponential.dist(1.0, shape=3)
chol, _, _ = pm.LKJCholeskyCov("chol_cov", n=3, eta=2, sd_dist=sd_dist, compute_corr=True)
ksi = pm.MvNormal("ksi", 0, chol=chol, dims=("obs", "latent"))
## Build Regression Components
coefs = pm.Normal('betas', 0, .5, dims='paths')
zeros = pt.zeros((3, 3))
## adjust ~ risk
zeros1 = pt.set_subtensor(zeros[1, 0], coefs[0])
## achieve ~ risk + adjust
zeros2 = pt.set_subtensor(zeros1[1, 2], coefs[1])
coefs_ = pt.set_subtensor(zeros2[0, 2], coefs[2])
structural_relations = pm.Deterministic('endogenous_structural_paths', pm.math.dot(ksi, coefs_))
# Construct Pseudo Observation matrix based on Factor Loadings
m1 = ksi[obs_idx, 0] * lambdas_1[0] + structural_relations[obs_idx, 0] #adjust
m2 = ksi[obs_idx, 0] * lambdas_1[1] + structural_relations[obs_idx, 0] #adjust
m3 = ksi[obs_idx, 0] * lambdas_1[2] + structural_relations[obs_idx, 0] #adjust
m4 = ksi[obs_idx, 1] * lambdas_2[0] + structural_relations[obs_idx, 1] #risk
m5 = ksi[obs_idx, 1] * lambdas_2[1] + structural_relations[obs_idx, 1] #risk
m6 = ksi[obs_idx, 1] * lambdas_2[2] + structural_relations[obs_idx, 1] #risk
m7 = ksi[obs_idx, 2] * lambdas_3[0] + structural_relations[obs_idx, 2] #achieve
m8 = ksi[obs_idx, 2] * lambdas_3[1] + structural_relations[obs_idx, 2] #achieve
m9 = ksi[obs_idx, 2] * lambdas_3[2] + structural_relations[obs_idx, 2] #achieve
mu = pm.Deterministic("mu", pm.math.stack([m1, m2, m3, m4, m5, m6, m7, m8, m9]).T)
## Error Terms
Psi = pm.InverseGamma("Psi", 2, 13, dims="indicators")
# Likelihood
_ = pm.Normal(
"likelihood",
mu,
Psi,
observed=df[coords['indicators']].values,
)
idata = pm.sample(
draws=2500,
chains=4,
#nuts_sampler="numpyro",
target_accept=0.90, idata_kwargs={"log_likelihood": True},
random_seed=150,
tune=1000,
)
idata.extend(pm.sample_posterior_predictive(idata))
return idata, model
idata_sem, model_sem = make_sem()Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [lambdas1_, lambdas2_, lambdas3_, chol_cov, ksi, betas, Psi]
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/multiprocessing/popen_fork.py:66: RuntimeWarning: os.fork() was called. os.fork() is incompatible with multithreaded code, and JAX is multithreaded, so this will likely lead to a deadlock.
self.pid = os.fork()/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/multiprocessing/popen_fork.py:66: RuntimeWarning: os.fork() was called. os.fork() is incompatible with multithreaded code, and JAX is multithreaded, so this will likely lead to a deadlock.
self.pid = os.fork()Sampling 4 chains for 1_000 tune and 2_500 draw iterations (4_000 + 10_000 draws total) took 170 seconds.
There were 1 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
Sampling: [likelihood]pm.model_to_graphviz(model_sem)az.summary(idata_sem, var_names=["lambdas1", "lambdas2", "lambdas3", "betas"])/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/diagnostics.py:596: RuntimeWarning: invalid value encountered in scalar divide
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| lambdas1[motiv] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 10000.0 | 10000.0 | NaN |
| lambdas1[harm] | 0.850 | 0.045 | 0.766 | 0.933 | 0.001 | 0.001 | 997.0 | 2361.0 | 1.00 |
| lambdas1[stabi] | 0.651 | 0.052 | 0.556 | 0.748 | 0.002 | 0.001 | 988.0 | 3122.0 | 1.00 |
| lambdas2[verbal] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 10000.0 | 10000.0 | NaN |
| lambdas2[ses] | 0.833 | 0.081 | 0.681 | 0.986 | 0.002 | 0.001 | 2282.0 | 4694.0 | 1.00 |
| lambdas2[ppsych] | -0.764 | 0.079 | -0.920 | -0.623 | 0.002 | 0.001 | 2399.0 | 4215.0 | 1.00 |
| lambdas3[read] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 10000.0 | 10000.0 | NaN |
| lambdas3[arith] | 0.610 | 0.057 | 0.498 | 0.712 | 0.003 | 0.002 | 381.0 | 1068.0 | 1.00 |
| lambdas3[spell] | 0.825 | 0.037 | 0.755 | 0.894 | 0.001 | 0.001 | 757.0 | 1717.0 | 1.00 |
| betas[risk->adjust] | 0.202 | 0.139 | -0.065 | 0.457 | 0.010 | 0.007 | 186.0 | 202.0 | 1.02 |
| betas[risk-->achieve] | 0.207 | 0.178 | -0.129 | 0.534 | 0.012 | 0.009 | 208.0 | 487.0 | 1.01 |
| betas[adjust-->achieve] | 0.747 | 0.104 | 0.556 | 0.942 | 0.006 | 0.004 | 311.0 | 810.0 | 1.01 |
make_ppc(idata_sem)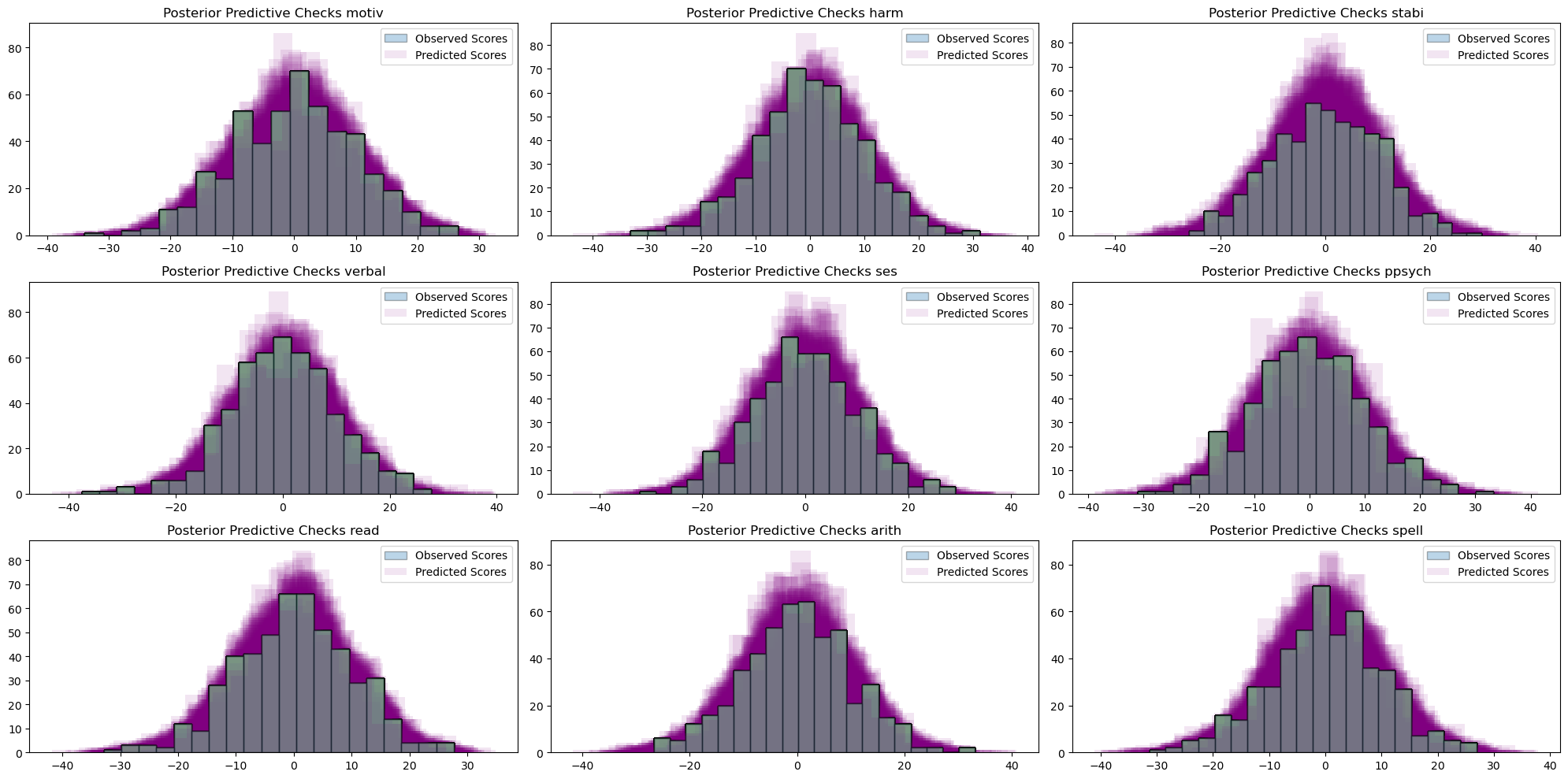
residuals_posterior_cov = get_posterior_resids(idata_sem, 1000)
residuals_posterior_corr = get_posterior_resids(idata_sem, 1000, metric="corr")fig, ax = plt.subplots(figsize=(20, 10))
mask = np.triu(np.ones_like(residuals_posterior_corr, dtype=bool))
ax = sns.heatmap(residuals_posterior_corr, annot=True, cmap="bwr", mask=mask)
ax.set_title("Residuals between Model Implied and Sample Correlations \n SEM model", fontsize=25);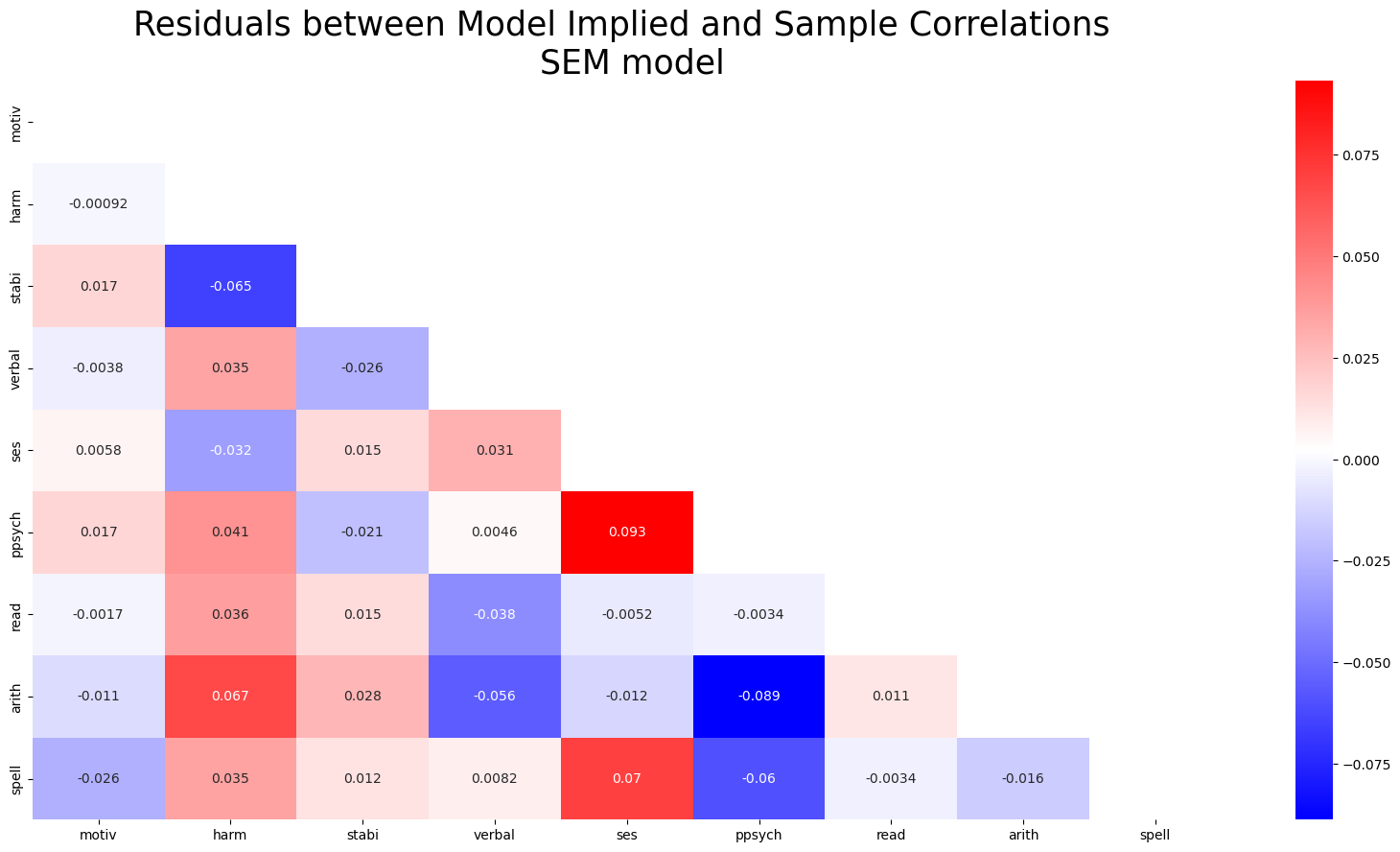
fig, axs = plt.subplots(1, 3, figsize=(15, 10))
axs = axs.flatten()
ax = axs[0]
ax1 = axs[1]
ax2 = axs[2]
az.plot_forest(idata_sem, var_names=["ksi"], combined=True, ax=ax, coords={"latent": ["adjust"]})
az.plot_forest(
idata_sem,
var_names=["ksi"],
combined=True,
ax=ax1,
colors="forestgreen",
coords={"latent": ["risk"]},
)
az.plot_forest(
idata_sem,
var_names=["ksi"],
combined=True,
ax=ax2,
colors="slateblue",
coords={"latent": ["achieve"]},
)
ax.set_yticklabels([])
ax.set_xlabel("ADJUST")
ax1.set_yticklabels([])
ax1.set_xlabel("RISK")
ax2.set_yticklabels([])
ax2.set_xlabel("ACHIEVE")
ax.axvline(-2, color="red")
ax1.axvline(-2, color="red")
ax2.axvline(-2, color="red")
ax.set_title("Individual On Latent Factor ADJUST")
ax1.set_title("Individual On Latent Factor RISK")
ax2.set_title("Individual On Latent Factor ACHIEVE")
plt.show();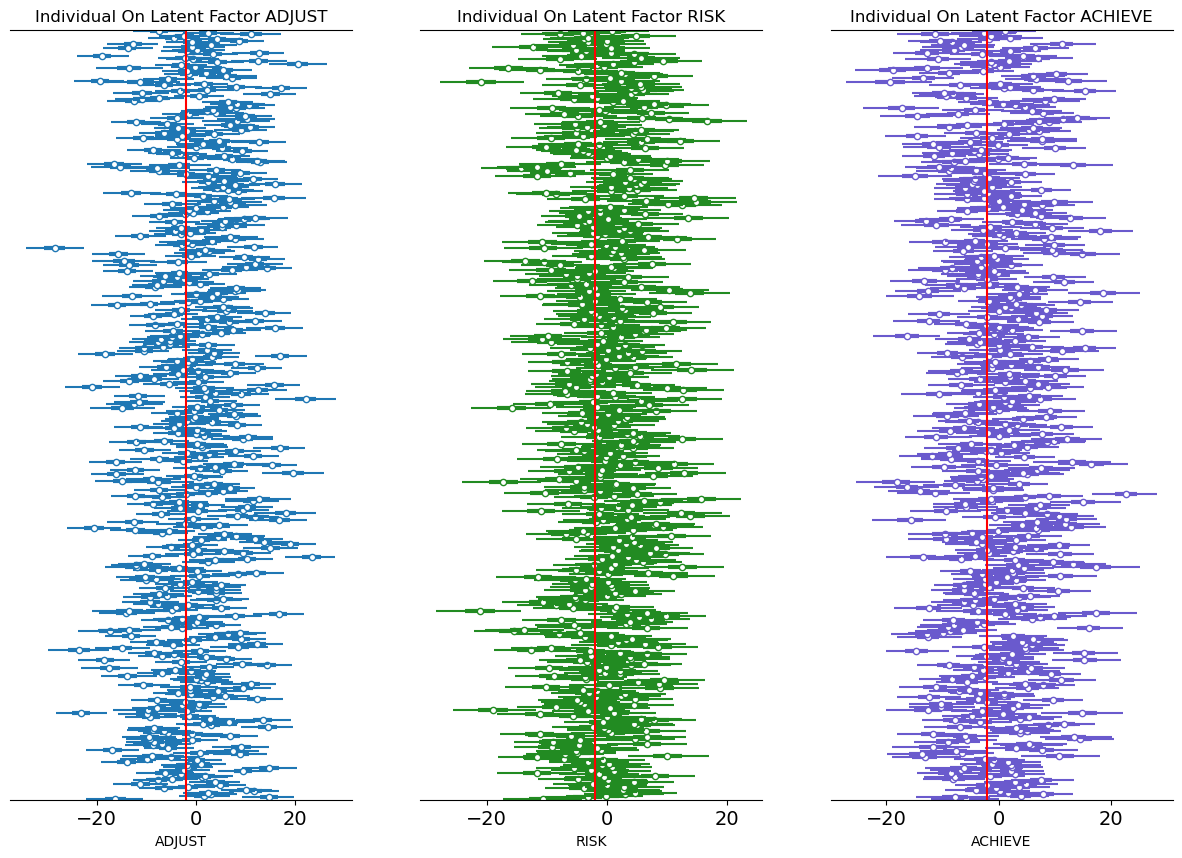
compare_df = az.compare({'SEM': idata_sem, 'CFA': idata}, 'waic')
compare_df/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/stats.py:1653: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(
/Users/nathanielforde/mambaforge/envs/bayesian_causal_book/lib/python3.10/site-packages/arviz/stats/stats.py:1653: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(| rank | elpd_waic | p_waic | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| SEM | 0 | -14525.202949 | 905.303983 | 0.000000 | 1.0 | 49.632750 | 0.000000 | True | log |
| CFA | 1 | -14748.968775 | 939.024032 | 223.765826 | 0.0 | 48.571539 | 12.080799 | True | log |
az.plot_compare(compare_df);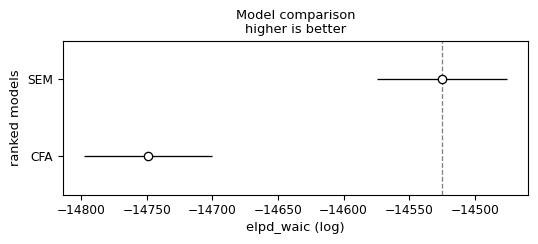
Counterfactual Analysis
model_sem\[ \begin{array}{rcl} \text{lambdas1\_} &\sim & \operatorname{Normal}(1,~0.5)\\\text{lambdas2\_} &\sim & \operatorname{Normal}(1,~0.5)\\\text{lambdas3\_} &\sim & \operatorname{Normal}(1,~0.5)\\\text{chol\_cov} &\sim & \operatorname{\_lkjcholeskycov}(3,~2,~\operatorname{Exponential}(f()))\\\text{ksi} &\sim & \operatorname{MultivariateNormal}(f(\text{chol\_cov}),~f(\text{chol\_cov}))\\\text{betas} &\sim & \operatorname{Normal}(0,~0.5)\\\text{Psi} &\sim & \operatorname{InverseGamma}(2,~13)\\\text{lambdas1} &\sim & \operatorname{Deterministic}(f(\text{lambdas1\_}))\\\text{lambdas2} &\sim & \operatorname{Deterministic}(f(\text{lambdas2\_}))\\\text{lambdas3} &\sim & \operatorname{Deterministic}(f(\text{lambdas3\_}))\\\text{chol\_cov\_corr} &\sim & \operatorname{Deterministic}(f(\text{chol\_cov}))\\\text{chol\_cov\_stds} &\sim & \operatorname{Deterministic}(f(\text{chol\_cov}))\\\text{endogenous\_structural\_paths} &\sim & \operatorname{Deterministic}(f(\text{ksi},~\text{betas}))\\\text{mu} &\sim & \operatorname{Deterministic}(f(\text{ksi},~\text{betas},~\text{lambdas3\_},~\text{lambdas2\_},~\text{lambdas1\_}))\\\text{likelihood} &\sim & \operatorname{Normal}(\text{mu},~\text{Psi}) \end{array} \]
model_beta0 = do(model_sem, {"betas": [0, 0 , 0]}, prune_vars=True)
model_beta1 = do(model_sem, {"betas": [.6, .3, .7]}, prune_vars=True)
model_beta0\[ \begin{array}{rcl} \text{lambdas1\_} &\sim & \operatorname{Normal}(1,~0.5)\\\text{lambdas2\_} &\sim & \operatorname{Normal}(1,~0.5)\\\text{lambdas3\_} &\sim & \operatorname{Normal}(1,~0.5)\\\text{chol\_cov} &\sim & \operatorname{\_lkjcholeskycov}(3,~2,~\operatorname{Exponential}(f()))\\\text{ksi} &\sim & \operatorname{MultivariateNormal}(f(\text{chol\_cov}),~f(\text{chol\_cov}))\\\text{Psi} &\sim & \operatorname{InverseGamma}(2,~13)\\\text{endogenous\_structural\_paths} &\sim & \operatorname{Deterministic}(f(\text{ksi}))\\\text{lambdas3} &\sim & \operatorname{Deterministic}(f(\text{lambdas3\_}))\\\text{lambdas2} &\sim & \operatorname{Deterministic}(f(\text{lambdas2\_}))\\\text{lambdas1} &\sim & \operatorname{Deterministic}(f(\text{lambdas1\_}))\\\text{mu} &\sim & \operatorname{Deterministic}(f(\text{ksi},~\text{lambdas3\_},~\text{lambdas2\_},~\text{lambdas1\_}))\\\text{likelihood} &\sim & \operatorname{Normal}(f(\text{ksi},~\text{lambdas1\_},~\text{lambdas2\_},~\text{lambdas3\_}),~\text{Psi}) \end{array} \]
pm.model_to_graphviz(model_beta0)# Sample new sales data assuming Google Ads off: P(Y | c, do(z=0))
idata_z0 = pm.sample_posterior_predictive(
idata_sem,
model=model_beta0,
predictions=False,
var_names=["likelihood", "betas",],
)
# Sample new sales data assuming Google Ads off: P(Y | c, do(z=1))
idata_z1 = pm.sample_posterior_predictive(
idata_sem,
model=model_beta1,
predictions=False,
var_names=["likelihood", "betas"],
)Sampling: [likelihood]Sampling: [likelihood]pd.DataFrame((idata_z0['posterior_predictive']['likelihood'] - idata_z1['posterior_predictive']['likelihood']).mean(dim=('chain', 'draw')).values)| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.463197 | -0.358005 | -0.286565 | -0.114874 | -0.042991 | 0.110666 | 4.936211 | 5.107738 | 4.972490 |
| 1 | 2.377638 | 2.422167 | 2.359539 | -0.063802 | -0.151001 | -0.018147 | -0.475168 | -0.303663 | -0.511428 |
| 2 | 4.663123 | 4.648260 | 4.735318 | 0.092329 | -0.014905 | -0.087927 | -5.440901 | -5.441826 | -5.468720 |
| 3 | -2.977008 | -2.997935 | -2.989136 | -0.039031 | 0.041482 | -0.025136 | 0.108632 | 0.264840 | 0.129751 |
| 4 | 1.826385 | 1.823625 | 1.722291 | 0.008998 | -0.087842 | 0.038139 | 3.982011 | 4.043272 | 3.869997 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 495 | -2.844905 | -3.038198 | -2.995460 | -0.007683 | 0.025422 | 0.177166 | -3.240432 | -3.093730 | -3.218599 |
| 496 | -1.817311 | -1.728041 | -1.782307 | 0.086392 | -0.032205 | -0.147835 | -3.111101 | -3.096323 | -3.237516 |
| 497 | 0.055163 | 0.142330 | 0.268756 | -0.165110 | -0.045748 | 0.036947 | -1.071530 | -1.102467 | -1.087500 |
| 498 | 1.989527 | 1.991247 | 1.913922 | -0.025677 | 0.027728 | -0.097815 | -9.445563 | -9.406408 | -9.387214 |
| 499 | 6.370377 | 6.422907 | 6.339601 | -0.023407 | -0.002096 | 0.105869 | 14.494246 | 14.581555 | 14.492164 |
500 rows × 9 columns
pd.DataFrame((idata_z0['posterior_predictive']['likelihood'] - idata_z1['posterior_predictive']['likelihood']).mean(dim=('chain', 'draw')).values).median()0 -0.105562
1 -0.085492
2 -0.135569
3 -0.006139
4 -0.004415
5 -0.002360
6 -0.148865
7 -0.163581
8 -0.169653
dtype: float64residuals_posterior_corr0 = get_posterior_resids(idata_z0, 1000, metric="corr")
residuals_posterior_corr1 = get_posterior_resids(idata_z1, 1000, metric="corr")residuals_posterior_corr0 - residuals_posterior_corr1| motiv | harm | stabi | verbal | ses | ppsych | read | arith | spell | |
|---|---|---|---|---|---|---|---|---|---|
| motiv | 0.000000 | -0.080177 | -0.131360 | -0.243116 | -0.192810 | 0.178007 | -0.899440 | -0.816758 | -0.882780 |
| harm | -0.080177 | 0.000000 | -0.153191 | -0.252675 | -0.206456 | 0.188974 | -0.799793 | -0.729367 | -0.786353 |
| stabi | -0.131360 | -0.153191 | 0.000000 | -0.262698 | -0.217575 | 0.197371 | -0.678641 | -0.611510 | -0.658578 |
| verbal | -0.243116 | -0.252675 | -0.262698 | 0.000000 | -0.010569 | -0.003814 | -0.241953 | -0.253102 | -0.250916 |
| ses | -0.192810 | -0.206456 | -0.217575 | -0.010569 | 0.000000 | 0.001399 | -0.196381 | -0.206370 | -0.204542 |
| ppsych | 0.178007 | 0.188974 | 0.197371 | -0.003814 | 0.001399 | 0.000000 | 0.175259 | 0.185912 | 0.186708 |
| read | -0.899440 | -0.799793 | -0.678641 | -0.241953 | -0.196381 | 0.175259 | 0.000000 | -0.178436 | -0.087744 |
| arith | -0.816758 | -0.729367 | -0.611510 | -0.253102 | -0.206370 | 0.185912 | -0.178436 | 0.000000 | -0.219602 |
| spell | -0.882780 | -0.786353 | -0.658578 | -0.250916 | -0.204542 | 0.186708 | -0.087744 | -0.219602 | 0.000000 |
fig, ax = plt.subplots(figsize=(20, 10))
mask = np.triu(np.ones_like(residuals_posterior_corr1, dtype=bool))
ax = sns.heatmap(residuals_posterior_corr1, annot=True, cmap="bwr", mask=mask, vmax=1, vmin=-1)
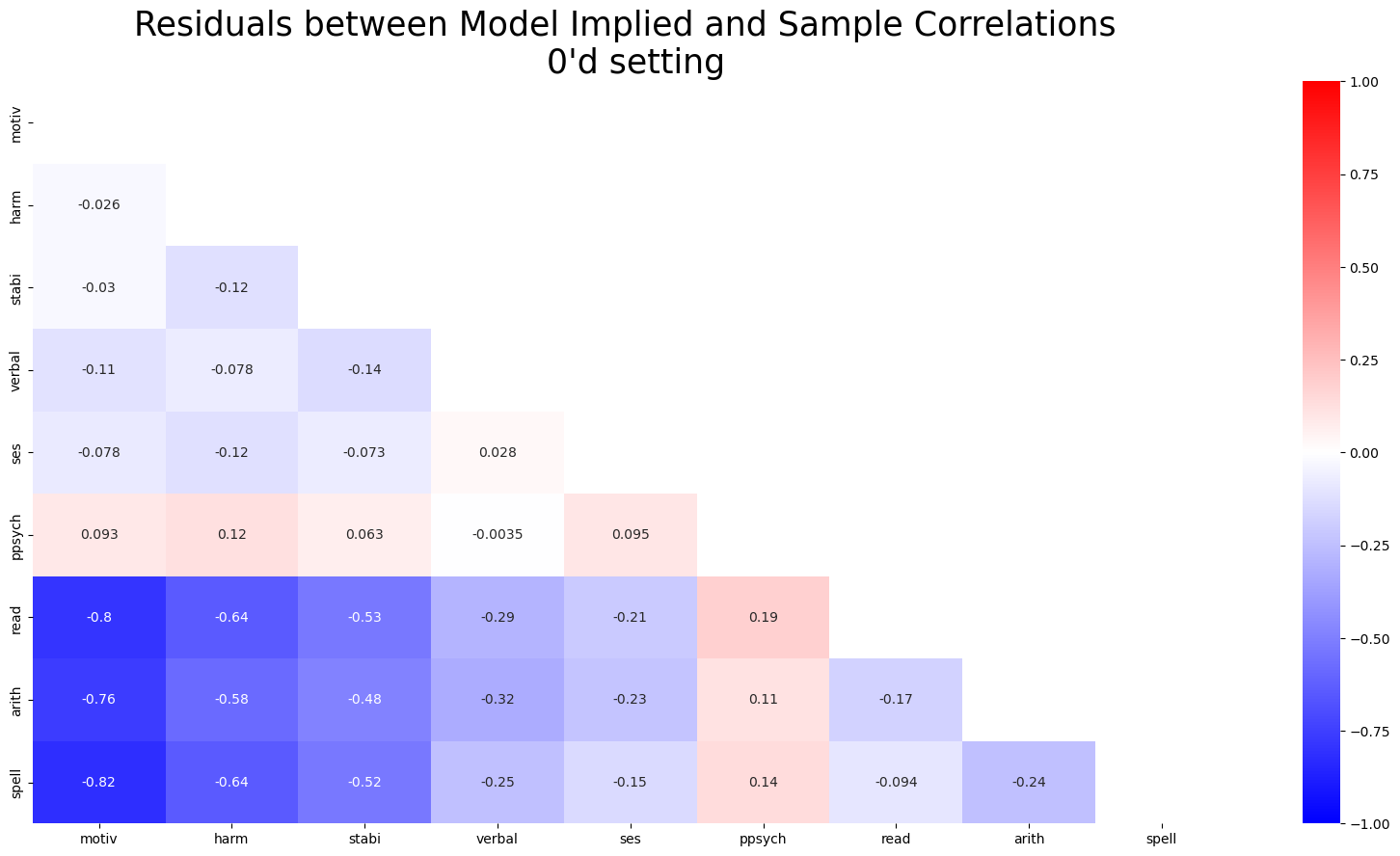
ax.set_title("Residuals between Model Implied and Sample Correlations \n Counterfactual Setting ", fontsize=25);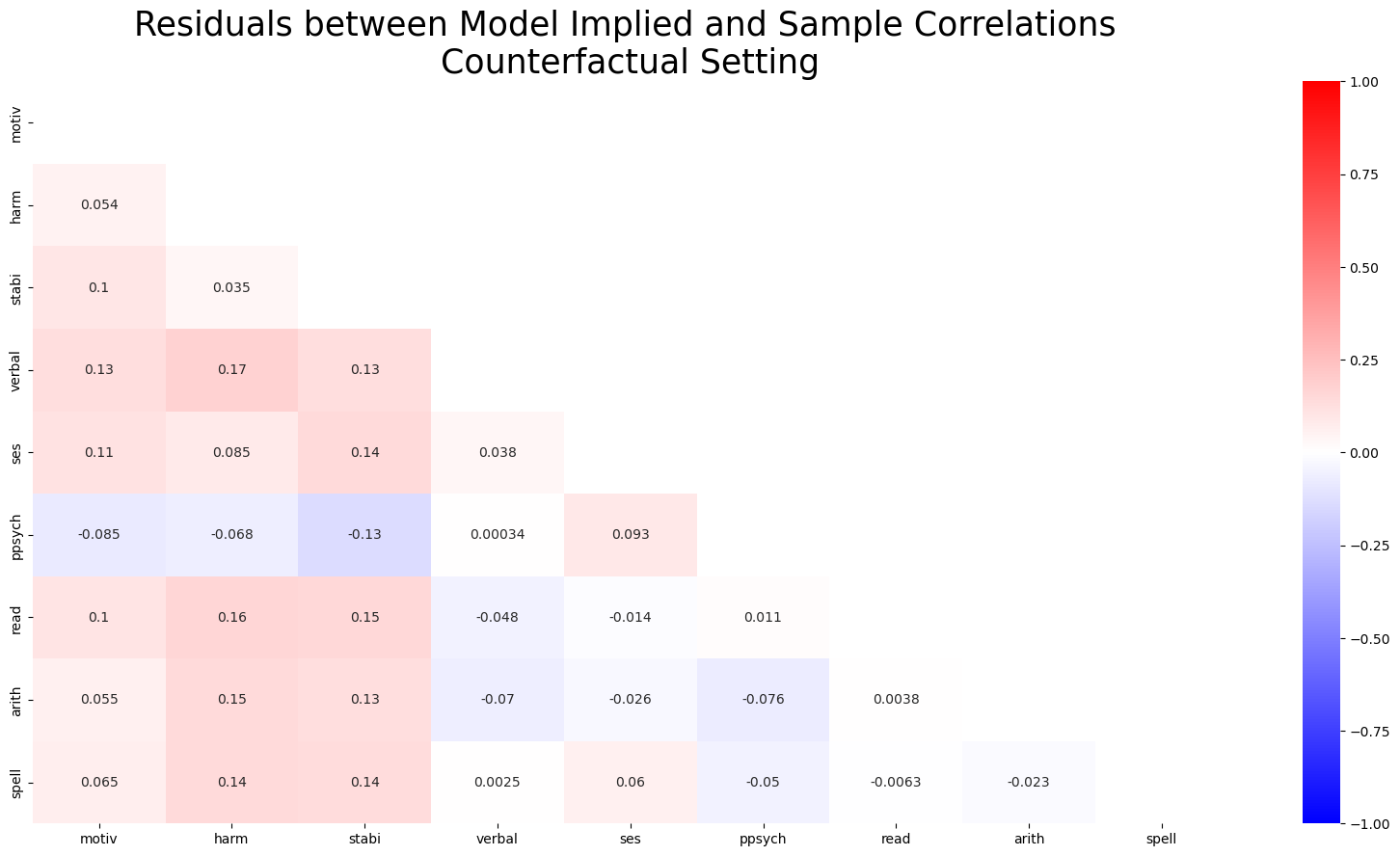
fig, ax = plt.subplots(figsize=(20, 10))
mask = np.triu(np.ones_like(residuals_posterior_corr0, dtype=bool))
ax = sns.heatmap(residuals_posterior_corr0, annot=True, cmap="bwr", mask=mask, vmax=1, vmin=-1)
ax.set_title("Residuals between Model Implied and Sample Correlations \n 0'd setting", fontsize=25);